

- #SNAPGENE PRIMER DESIGN VERIFICATION#
- #SNAPGENE PRIMER DESIGN SOFTWARE#
- #SNAPGENE PRIMER DESIGN LICENSE#
- #SNAPGENE PRIMER DESIGN MAC#
#SNAPGENE PRIMER DESIGN VERIFICATION#
#SNAPGENE PRIMER DESIGN SOFTWARE#
Ok, so this is not exactly a plasmid mapping or DNA annotation tool, but this free software is quite comprehensive, and looks pretty cool. The software allows the user to save and export files in open standard formats (FASTA, Genbank, UniProt, etc) and has an easy to navigate Sequence Feature viewer. Along with restriction enzyme analysis, it has primer picking capabilities and a feature that allows you to analyze your oligos to determine melting temperatures, hairpin structures, or dimers. GenBeans is a free molecular biology program for DNA analysis developed by GeneInfinity. GenBeans is software written in Java to work on Windows, Linux, and Mac. Snapgene is compatible with file formats of many other popular programs such as ApE, Lasergene, Genbank, Vector NTI etc and can import not only the DNA sequences but the annotations, too.
#SNAPGENE PRIMER DESIGN LICENSE#
The licensed version ($345 for a single academic annual license or $695 or a single academic permanent license) provides additional perks such as sequence editing, restriction digest result predictions, and cloning simulations (PCR, Restriction Digests, Gibson, etc). There are 2 different products: The free SnapGene viewer can make maps and design primers. It combines a lot of great features with a good user interface that allows scientists to plan, visualize, and document cloning and PCR projects. SnapGene is versatile software available for Windows and OS X. It has the basic features a molecular biologist is looking for: annotation of sequence, primer and restriction enzyme search, graphic maps, alignments (between two sequences or a sequence and a trace file), and virtual digests. The program can import DNA Strider, FASTA, Genbank, EMBL and ABI files and exports data in DNA Strider-compatible or Genbank file formats. Updated in fall 2012, ApE is free plasmid editor that runs on all three major platforms: Windows, OS X, and Linux. For $300 you can purchase a licensed version that eliminates the requirement of regular updating, but the program features are pretty much the same. It automatically finds restriction sites, methylation sites (dam and dcm), ORFs, and potential primers. pDRAW32 is a basic, yet comprehensive, DNA analysis program that can be used for virtual cloning, annotations, and analysis.
#SNAPGENE PRIMER DESIGN MAC#
Recently updated, pDRAW32 is a free Windows program (but it may be run on a Mac with a porting software such as Wineskin).
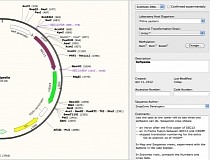
This includes two tried and true favorites, as well as some newer ones that may not have made it onto everyone’s radar yet. To help get you out of a molecular biology rut, I’m going to introduce you to a few DNA annotation/plasmid mapping programs. Sometimes though, we don’t want to spend the extra time learning a new program even though it might make our lives easier in the end. Don’t get me wrong-this is not necessarily a bad thing! Many times we don’t go looking for new software because the old stuff suits our needs. If you are like most molecular biologists, you probably use the same software your colleagues do-usually it is either the stuff that gets passed down from grad student to grad student or the one licensed software your lab pays for. A quick Google search brings up dozens of hits – but how do you know which one to use? Plasmid mapping and DNA annotation software is pretty abundant these days.


 0 kommentar(er)
0 kommentar(er)
